Phylogeny and Reconstructing Phylogenetic Trees
Introduction
So, what is phylogeny? It's the study of the evolution of life forms.
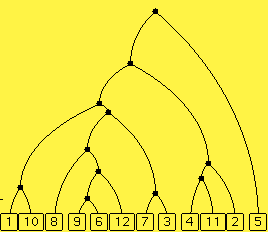
One phylogenetic tree, also called a cladogram or a
dendrogram, is displayed below. It
is a tree of several
life forms and their relations. In this display, time is the vertical
dimension with the current time at the bottom and earlier times above it.
There are ten extant species (species currently living) named from 1 through
10. The lines above the extant species represent the same species, just in
the past. When two lines converge to a point, that should be interpreted
as the point when the two species diverged from a common ancestral species,
the point being the common ancestral species. And so it goes until eventually,
some time in the past, all the species derived from just one species, the
one displayed as the top point.
The phylogenetic tree displayed above was randomly generated. If you like,
you can click on the button labelled "New Phyltree" to generate a different
phylogenetic tree. You can also change the number of extant species by editing
the field labelled "Species". (Press return after you change the number.)
A couple of comments about the horizontal dimension of the phylogenetic
tree. It doesn't mean anything! Any tree can be displayed in two dimensions
so that the lines don't cross,
and so these phylogenetic trees are. It is completely arbitrary whether a
branch of the tree is placed to the left or to the right of a dot. So, a tree
with two species, 1 and 2, with a common ancestor 3, could be drawn with
either species 1 or species 2 on the left, and the other species on the right.
About the random creation of the trees. The trees are randomly created
with n leaves (the number of current species). There are many possible algorithms
for such a construction. Here, a fairly random-looking tree is needed.
The algorithm is not based on any actual data or theory of species
evolution. The tree is created as follows. First, there are n
unrelated species. Usually, two are selected at random to have a common
ancestor, but 1/10 of the time, three are selected (and 1/100 of the
time four, etc.). An ancestral species is created for these
species, and it is placed back in time before the older of the child species.
The amount further back in time is 2/10 plus a random number between 0 and 1, which
makes an average of 0.7 time units back. This process is
repeated until all the species have a common ancestor.
The vertical scale shown is arbitrary, except that it is the same scale used
to generate the random tree.
Validity of the tree model. A tree isn't always the best model. Here are
some times when it isn't best.
- For individuals within a species. The genetic material of an individual
doesn't derive from a
single earlier existing individual. Animals and plants that multiply by sexual
reproduction receive half their genetic material from each of two parents, so a
tree like this is inappropriate. For
species that multiply asexually, a tree is appropriate. Even for species
that usually multiply asexually—such as many one-celled creatures—the occasional
exchange of genetic material through
conjugation is so important that trees are inappropriate.
- For closely related species. Individuals do occasionally mate between closely related species, and their progeny survive to contribute to the gene pool of one or both of the parent species.
As the species diverge, such intermixing of genetic material becomes more rare. One solution is to treat closely related species as one larger variable species. Another is simply not to consider
closely related species.
- Hybrid species. In the plant world it occasionally happens that a new tetraploid species arises from two diploid species. The two parent species need to be somewhat related for this to
happen.
- Distant interaction. There are a couple of ways that genetic material from one species can find its way into a distantly related or unrelated species. Among bacteria, sometimes a bacterium
of one species can ingest the genetic material of a bacterium of another species and incorporate part of it into its own genetic material. Rare as this may be, the effects are significant.
Sometimes viruses can inadvertantly transport genetic material from one species to another. When some viruses break out of cells of one species, they may infect other species and carry that material
to them.
In spite of these exceptions, a tree model is usually a pretty good model to show the relations among species.
Next page: mutations.
Table of contents:
http://aleph0.clarku.edu/~djoyce/java/Phyltree.
David E. Joyce
Department of Mathematics and Computer Science
Clark University
January, 1996